Regenerative medicine has come a long way, baby

After a cloned baby sheep, what started as one of the most controversial areas in medicine is now promising to transform it.
The field of regenerative medicine had a shaky start. In 2002, when news spread about the first cloned animal, Dolly the sheep, a raucous debate ensued. Scary headlines and organized opposition groups put pressure on government leaders, who responded by tightening restrictions on this type of research.
Fast forward to today, and regenerative medicine, which focuses on making unhealthy tissues and organs healthy again, is rewriting the code to healing many disorders, though it’s still young enough to be considered nascent. What started as one of the most controversial areas in medicine is now promising to transform it.
Progress in the lab has addressed previous concerns. Back in the early 2000s, some of the most fervent controversy centered around somatic cell nuclear transfer (SCNT), the process used by scientists to produce Dolly. There was fear that this technique could be used in humans, with possibly adverse effects, considering the many medical problems of the animals who had been cloned.
But today, scientists have discovered better approaches with fewer risks. Pioneers in the field are embracing new possibilities for cellular reprogramming, 3D organ printing, AI collaboration, and even growing organs in space. It could bring a new era of personalized medicine for longer, healthier lives - while potentially sparking new controversies.
Engineering tissues from amniotic fluids
Work in regenerative medicine seeks to reverse damage to organs and tissues by culling, modifying and replacing cells in the human body. Scientists in this field reach deep into the mechanisms of diseases and the breakdowns of cells, the little workhorses that perform all life-giving processes. If cells can’t do their jobs, they take whole organs and systems down with them. Regenerative medicine seeks to harness the power of healthy cells derived from stem cells to do the work that can literally restore patients to a state of health—by giving them healthy, functioning tissues and organs.
Modern-day regenerative medicine takes its origin from the 1998 isolation of human embryonic stem cells, first achieved by John Gearhart at Johns Hopkins University. Gearhart isolated the pluripotent cells that can differentiate into virtually every kind of cell in the human body. There was a raging controversy about the use of these cells in research because at that time they came exclusively from early-stage embryos or fetal tissue.
Back then, the highly controversial SCNT cells were the only way to produce genetically matched stem cells to treat patients. Since then, the picture has changed radically because other sources of highly versatile stem cells have been developed. Today, scientists can derive stem cells from amniotic fluid or reprogram patients’ skin cells back to an immature state, so they can differentiate into whatever types of cells the patient needs.
In the context of medical history, the field of regenerative medicine is progressing at a dizzying speed. But for those living with aggressive or chronic illnesses, it can seem that the wheels of medical progress grind slowly.
The ethical debate has been dialed back and, in the last few decades, the field has produced important innovations, spurring the development of whole new FDA processes and categories, says Anthony Atala, a bioengineer and director of the Wake Forest Institute for Regenerative Medicine. Atala and a large team of researchers have pioneered many of the first applications of 3D printed tissues and organs using cells developed from patients or those obtained from amniotic fluid or placentas.
His lab, considered to be the largest devoted to translational regenerative medicine, is currently working with 40 different engineered human tissues. Sixteen of them have been transplanted into patients. That includes skin, bladders, urethras, muscles, kidneys and vaginal organs, to name just a few.
These achievements are made possible by converging disciplines and technologies, such as cell therapies, bioengineering, gene editing, nanotechnology and 3D printing, to create living tissues and organs for human transplants. Atala is currently overseeing clinical trials to test the safety of tissues and organs engineered in the Wake Forest lab, a significant step toward FDA approval.
In the context of medical history, the field of regenerative medicine is progressing at a dizzying speed. But for those living with aggressive or chronic illnesses, it can seem that the wheels of medical progress grind slowly.
“It’s never fast enough,” Atala says. “We want to get new treatments into the clinic faster, but the reality is that you have to dot all your i’s and cross all your t’s—and rightly so, for the sake of patient safety. People want predictions, but you can never predict how much work it will take to go from conceptualization to utilization.”
As a surgeon, he also treats patients and is able to follow transplant recipients. “At the end of the day, the goal is to get these technologies into patients, and working with the patients is a very rewarding experience,” he says. Will the 3D printed organs ever outrun the shortage of donated organs? “That’s the hope,” Atala says, “but this technology won’t eliminate the need for them in our lifetime.”
New methods are out of this world
Jeanne Loring, another pioneer in the field and director of the Center for Regenerative Medicine at Scripps Research Institute in San Diego, says that investment in regenerative medicine is not only paying off, but is leading to truly personalized medicine, one of the holy grails of modern science.
This is because a patient’s own skin cells can be reprogrammed to become replacements for various malfunctioning cells causing incurable diseases, such as diabetes, heart disease, macular degeneration and Parkinson’s. If the cells are obtained from a source other than the patient, they can be rejected by the immune system. This means that patients need lifelong immunosuppression, which isn’t ideal. “With Covid,” says Loring, “I became acutely aware of the dangers of immunosuppression.” Using the patient’s own cells eliminates that problem.
Microgravity conditions make it easier for the cells to form three-dimensional structures, which could more easily lead to the growing of whole organs. In fact, Loring's own cells have been sent to the ISS for study.
Loring has a special interest in neurons, or brain cells that can be developed by manipulating cells found in the skin. She is looking to eventually treat Parkinson’s disease using them. The manipulated cells produce dopamine, the critical hormone or neurotransmitter lacking in the brains of patients. A company she founded plans to start a Phase I clinical trial using cell therapies for Parkinson’s soon, she says.
This is the culmination of many years of basic research on her part, some of it on her own cells. In 2007, Loring had her own cells reprogrammed, so there’s a cell line that carries her DNA. “They’re just like embryonic stem cells, but personal,” she said.
Loring has another special interest—sending immature cells into space to be studied at the International Space Station. There, microgravity conditions make it easier for the cells to form three-dimensional structures, which could more easily lead to the growing of whole organs. In fact, her own cells have been sent to the ISS for study. “My colleagues and I have completed four missions at the space station,” she says. “The last cells came down last August. They were my own cells reprogrammed into pluripotent cells in 2009. No one else can say that,” she adds.
Future controversies and tipping points
Although the original SCNT debate has calmed down, more controversies may arise, Loring thinks.
One of them could concern growing synthetic embryos. The embryos are ultimately derived from embryonic stem cells, and it’s not clear to what stage these embryos can or will be grown in an artificial uterus—another recent invention. The science, so far done only in animals, is still new and has not been widely publicized but, eventually, “People will notice the production of synthetic embryos and growing them in an artificial uterus,” Loring says. It’s likely to incite many of the same reactions as the use of embryonic stem cells.
Bernard Siegel, the founder and director of the Regenerative Medicine Foundation and executive director of the newly formed Healthspan Action Coalition (HSAC), believes that stem cell science is rapidly approaching tipping point and changing all of medical science. (For disclosure, I do consulting work for HSAC). Siegel says that regenerative medicine has become a new pillar of medicine that has recently been fast-tracked by new technology.
Artificial intelligence is speeding up discoveries and the convergence of key disciplines, as demonstrated in Atala’s lab, which is creating complex new medical products that replace the body’s natural parts. Just as importantly, those parts are genetically matched and pose no risk of rejection.
These new technologies must be regulated, which can be a challenge, Siegel notes. “Cell therapies represent a challenge to the existing regulatory structure, including payment, reimbursement and infrastructure issues that 20 years ago, didn’t exist.” Now the FDA and other agencies are faced with this revolution, and they’re just beginning to adapt.
Siegel cited the 2021 FDA Modernization Act as a major step. The Act allows drug developers to use alternatives to animal testing in investigating the safety and efficacy of new compounds, loosening the agency’s requirement for extensive animal testing before a new drug can move into clinical trials. The Act is a recognition of the profound effect that cultured human cells are having on research. Being able to test drugs using actual human cells promises to be far safer and more accurate in predicting how they will act in the human body, and could accelerate drug development.
Siegel, a longtime veteran and founding father of several health advocacy organizations, believes this work helped bring cell therapies to people sooner rather than later. His new focus, through the HSAC, is to leverage regenerative medicine into extending not just the lifespan but the worldwide human healthspan, the period of life lived with health and vigor. “When you look at the HSAC as a tree,” asks Siegel, “what are the roots of that tree? Stem cell science and the huge ecosystem it has created.” The study of human aging is another root to the tree that has potential to lengthen healthspans.
The revolutionary science underlying the extension of the healthspan needs to be available to the whole world, Siegel says. “We need to take all these roots and come up with a way to improve the life of all mankind,” he says. “Everyone should be able to take advantage of this promising new world.”
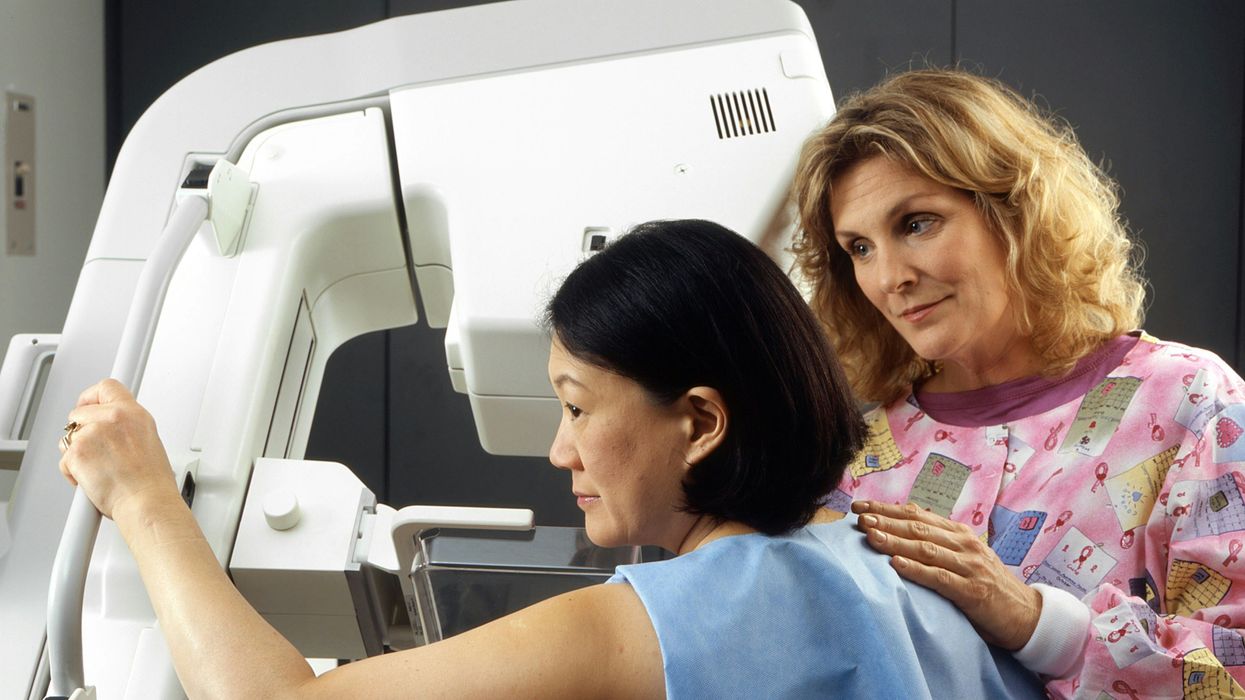
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.
Urinary tract infections account for more than 8 million trips to the doctor each year.
Few things are more painful than a urinary tract infection (UTI). Common in men and women, these infections account for more than 8 million trips to the doctor each year and can cause an array of uncomfortable symptoms, from a burning feeling during urination to fever, vomiting, and chills. For an unlucky few, UTIs can be chronic—meaning that, despite treatment, they just keep coming back.
But new research, presented at the European Association of Urology (EAU) Congress in Paris this week, brings some hope to people who suffer from UTIs.
Clinicians from the Royal Berkshire Hospital presented the results of a long-term, nine-year clinical trial where 89 men and women who suffered from recurrent UTIs were given an oral vaccine called MV140, designed to prevent the infections. Every day for three months, the participants were given two sprays of the vaccine (flavored to taste like pineapple) and then followed over the course of nine years. Clinicians analyzed medical records and asked the study participants about symptoms to check whether any experienced UTIs or had any adverse reactions from taking the vaccine.
The results showed that across nine years, 48 of the participants (about 54%) remained completely infection-free. On average, the study participants remained infection free for 54.7 months—four and a half years.
“While we need to be pragmatic, this vaccine is a potential breakthrough in preventing UTIs and could offer a safe and effective alternative to conventional treatments,” said Gernot Bonita, Professor of Urology at the Alta Bro Medical Centre for Urology in Switzerland, who is also the EAU Chairman of Guidelines on Urological Infections.
The news comes as a relief not only for people who suffer chronic UTIs, but also to doctors who have seen an uptick in antibiotic-resistant UTIs in the past several years. Because UTIs usually require antibiotics, patients run the risk of developing a resistance to the antibiotics, making infections more difficult to treat. A preventative vaccine could mean less infections, less antibiotics, and less drug resistance overall.
“Many of our participants told us that having the vaccine restored their quality of life,” said Dr. Bob Yang, Consultant Urologist at the Royal Berkshire NHS Foundation Trust, who helped lead the research. “While we’re yet to look at the effect of this vaccine in different patient groups, this follow-up data suggests it could be a game-changer for UTI prevention if it’s offered widely, reducing the need for antibiotic treatments.”