Her Incredible Sense of Smell Helped Scientists Develop the First Parkinson's Test

Joy Milne's unusual sense of smell led Dr. Tilo Kunath, a neurobiologist at the Centre for Regenerative Medicine at the University of Edinburgh, and a host of other scientists, to develop a new diagnostic test for Parkinson's.
Forty years ago, Joy Milne, a nurse from Perth, Scotland, noticed a musky odor coming from her husband, Les. At first, Milne thought the smell was a result of bad hygiene and badgered her husband to take longer showers. But when the smell persisted, Milne learned to live with it, not wanting to hurt her husband's feelings.
Twelve years after she first noticed the "woodsy" smell, Les was diagnosed at the age of 44 with Parkinson's Disease, a neurodegenerative condition characterized by lack of dopamine production and loss of movement. Parkinson's Disease currently affects more than 10 million people worldwide.
Milne spent the next several years believing the strange smell was exclusive to her husband. But to her surprise, at a local support group meeting in 2012, she caught the familiar scent once again, hanging over the group like a cloud. Stunned, Milne started to wonder if the smell was the result of Parkinson's Disease itself.
Milne's discovery led her to Dr. Tilo Kunath, a neurobiologist at the Centre for Regenerative Medicine at the University of Edinburgh. Together, Milne, Kunath, and a host of other scientists would use Milne's unusual sense of smell to develop a new diagnostic test, now in development and poised to revolutionize the treatment of Parkinson's Disease.
"Joy was in the audience during a talk I was giving on my work, which has to do with Parkinson's and stem cell biology," Kunath says. "During the patient engagement portion of the talk, she asked me if Parkinson's had a smell to it." Confused, Kunath said he had never heard of this – but for months after his talk he continued to turn the question over in his mind.
Kunath knew from his research that the skin's microbiome changes during different disease processes, releasing metabolites that can give off odors. In the medical literature, diseases like melanoma and Type 2 diabetes have been known to carry a specific scent – but no such connection had been made with Parkinson's. If people could smell Parkinson's, he thought, then it stood to reason that those metabolites could be isolated, identified, and used to potentially diagnose Parkinson's by their presence alone.
First, Kunath and his colleagues decided to test Milne's sense of smell. "I got in touch with Joy again and we designed a protocol to test her sense of smell without her having to be around patients," says Kunath, which could have affected the validity of the test. In his spare time, Kunath collected t-shirt samples from people diagnosed with Parkinson's and from others without the diagnosis and gave them to Milne to smell. In 100 percent of the samples, Milne was able to detect whether a person had Parkinson's based on smell alone. Amazingly, Milne was even able to detect the "Parkinson's scent" in a shirt from the control group – someone who did not have a Parkinson's diagnosis, but would go on to be diagnosed nine months later.
From the initial study, the team discovered that Parkinson's did have a smell, that Milne – inexplicably – could detect it, and that she could detect it long before diagnosis like she had with her husband, Les. But the experiments revealed other things that the team hadn't been expecting.
"One surprising thing we learned from that experiment was that the odor was always located in the back of the shirt – never in the armpit, where we expected the smell to be," Kunath says. "I had a chance meeting with a dermatologist and he said the smell was due to the patient's sebum, which are greasy secretions that are really dense on your upper back. We have sweat glands, instead of sebum, in our armpits." Patients with Parkinson's are also known to have increased sebum production.
With the knowledge that a patient's sebum was the source of the unusual smell, researchers could go on to investigate exactly what metabolites were in the sebum and in what amounts. Kunath, along with his associate, Dr. Perdita Barran, collected and analyzed sebum samples from 64 participants across the United Kingdom. Once the samples were collected, Barran and others analyzed it using a method called gas chromatography mass spectrometry, or GS-MC, which separated, weighed and helped identify the individual compounds present in each sebum sample.
Barran's team can now correctly identify Parkinson's in nine out of 10 patients – a much quicker and more accurate way to diagnose than what clinicians do now.
"The compounds we've identified in the sebum are not unique to people with Parkinson's, but they are differently expressed," says Barran, a professor of mass spectrometry at the University of Manchester. "So this test we're developing now is not a black-and-white, do-you-have-something kind of test, but rather how much of these compounds do you have compared to other people and other compounds." The team identified over a dozen compounds that were present in the sebum of Parkinson's patients in much larger amounts than the control group.
Using only the GC-MS and a sebum swab test, Barran's team can now correctly identify Parkinson's in nine out of 10 patients – a much quicker and more accurate way to diagnose than what clinicians do now.
"At the moment, a clinical diagnosis is based on the patient's physical symptoms," Barran says, and determining whether a patient has Parkinson's is often a long and drawn-out process of elimination. "Doctors might say that a group of symptoms looks like Parkinson's, but there are other reasons people might have those symptoms, and it might take another year before they're certain," Barran says. "Some of those symptoms are just signs of aging, and other symptoms like tremor are present in recovering alcoholics or people with other kinds of dementia." People under the age of 40 with Parkinson's symptoms, who present with stiff arms, are often misdiagnosed with carpal tunnel syndrome, she adds.
Additionally, by the time physical symptoms are present, Parkinson's patients have already lost a substantial amount of dopamine receptors – about sixty percent -- in the brain's basal ganglia. Getting a diagnosis before physical symptoms appear would mean earlier interventions that could prevent dopamine loss and preserve regular movement, Barran says.
"Early diagnosis is good if it means there's a chance of early intervention," says Barran. "It stops the process of dopamine loss, which means that motor symptoms potentially will not happen, or the onset of symptoms will be substantially delayed." Barran's team is in the processing of streamlining the sebum test so that definitive results will be ready in just two minutes.
"What we're doing right now will be a very inexpensive test, a rapid-screen test, and that will encourage people to self-sample and test at home," says Barran. In addition to diagnosing Parkinson's, she says, this test could also be potentially useful to determine if medications were at a therapeutic dose in people who have the disease, since the odor is strongest in people whose symptoms are least controlled by medication.
"When symptoms are under control, the odor is lower," Barran says. "Potentially this would allow patients and clinicians to see whether their symptoms are being managed properly with medication, or perhaps if they're being overmedicated." Hypothetically, patients could also use the test to determine if interventions like diet and exercise are effective at keeping Parkinson's controlled.
"We hope within the next two to five years we will have a test available."
Barran is now running another clinical trial – one that determines whether they can diagnose at an earlier stage and whether they can identify a difference in sebum samples between different forms of Parkinson's or diseases that have Parkinson's-like symptoms, such as Lewy Body Dementia.
"Within the next one to two years, we hope to be running a trial in the Manchester area for those people who do not have motor symptoms but are at risk for developing dementia due to symptoms like loss of smell and sleep difficulty," Barran had said in 2019. "If we can establish that, we can roll out a test that determines if you have Parkinson's or not with those first pre-motor symptoms, and then at what stage. We hope within the next two to five years we will have a test available."
In a 2022 study, published in the American Chemical Society, researchers used mass spectrometry to analyze sebum from skin swabs for the presence of the specific molecules. They found that some specific molecules are present only in people who have Parkinson’s. Now they hope that the same method can be used in regular diagnostic labs. The test, many years in the making, is inching its way to the clinic.
"We would likely first give this test to people who are at risk due to a genetic predisposition, or who are at risk based on prodomal symptoms, like people who suffer from a REM sleep disorder who have a 50 to 70 percent chance of developing Parkinson's within a ten year period," Barran says. "Those would be people who would benefit from early therapeutic intervention. For the normal population, it isn't beneficial at the moment to know until we have therapeutic interventions that can be useful."
Milne's husband, Les, passed away from complications of Parkinson's Disease in 2015. But thanks to him and the dedication of his wife, Joy, science may have found a way to someday prolong the lives of others with this devastating disease. Sometimes she can smell people who have Parkinson’s while in the supermarket or walking down the street but has been told by medical ethicists she cannot tell them, Milne said in an interview with the Guardian. But once the test becomes available in the clinics, it will do the job for her.
[Ed. Note: A older version of this hit article originally ran on September 3, 2019.]
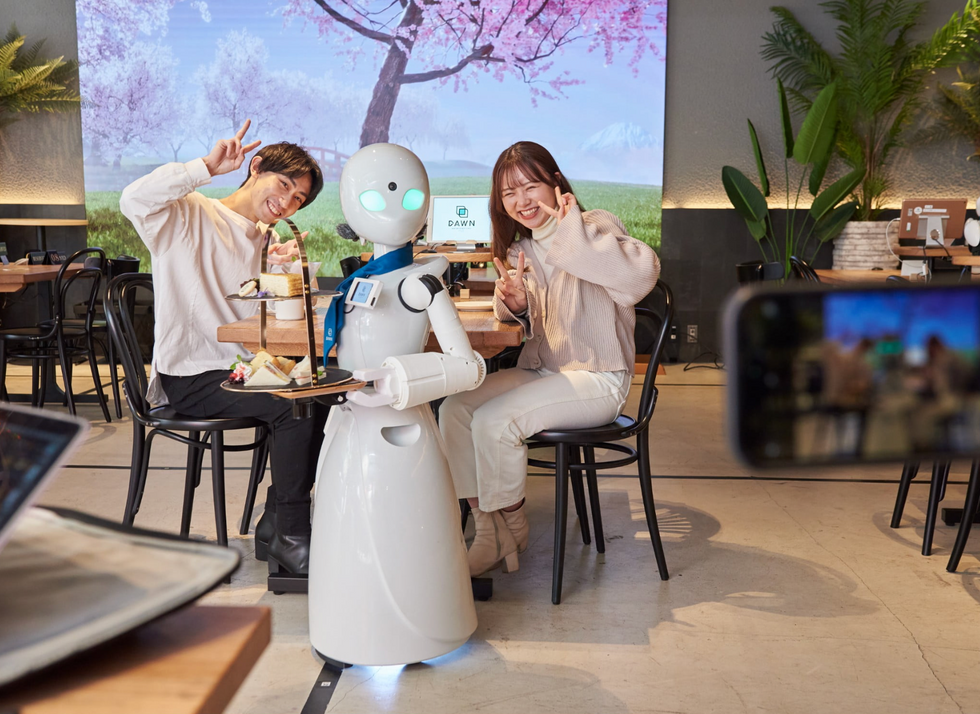
A robot server, controlled remotely by a disabled worker, delivers drinks to patrons at the DAWN cafe in Tokyo.
A sleek, four-foot tall white robot glides across a cafe storefront in Tokyo’s Nihonbashi district, holding a two-tiered serving tray full of tea sandwiches and pastries. The cafe’s patrons smile and say thanks as they take the tray—but it’s not the robot they’re thanking. Instead, the patrons are talking to the person controlling the robot—a restaurant employee who operates the avatar from the comfort of their home.
It’s a typical scene at DAWN, short for Diverse Avatar Working Network—a cafe that launched in Tokyo six years ago as an experimental pop-up and quickly became an overnight success. Today, the cafe is a permanent fixture in Nihonbashi, staffing roughly 60 remote workers who control the robots remotely and communicate to customers via a built-in microphone.
More than just a creative idea, however, DAWN is being hailed as a life-changing opportunity. The workers who control the robots remotely (known as “pilots”) all have disabilities that limit their ability to move around freely and travel outside their homes. Worldwide, an estimated 16 percent of the global population lives with a significant disability—and according to the World Health Organization, these disabilities give rise to other problems, such as exclusion from education, unemployment, and poverty.
These are all problems that Kentaro Yoshifuji, founder and CEO of Ory Laboratory, which supplies the robot servers at DAWN, is looking to correct. Yoshifuji, who was bedridden for several years in high school due to an undisclosed health problem, launched the company to help enable people who are house-bound or bedridden to more fully participate in society, as well as end the loneliness, isolation, and feelings of worthlessness that can sometimes go hand-in-hand with being disabled.
“It’s heartbreaking to think that [people with disabilities] feel they are a burden to society, or that they fear their families suffer by caring for them,” said Yoshifuji in an interview in 2020. “We are dedicating ourselves to providing workable, technology-based solutions. That is our purpose.”
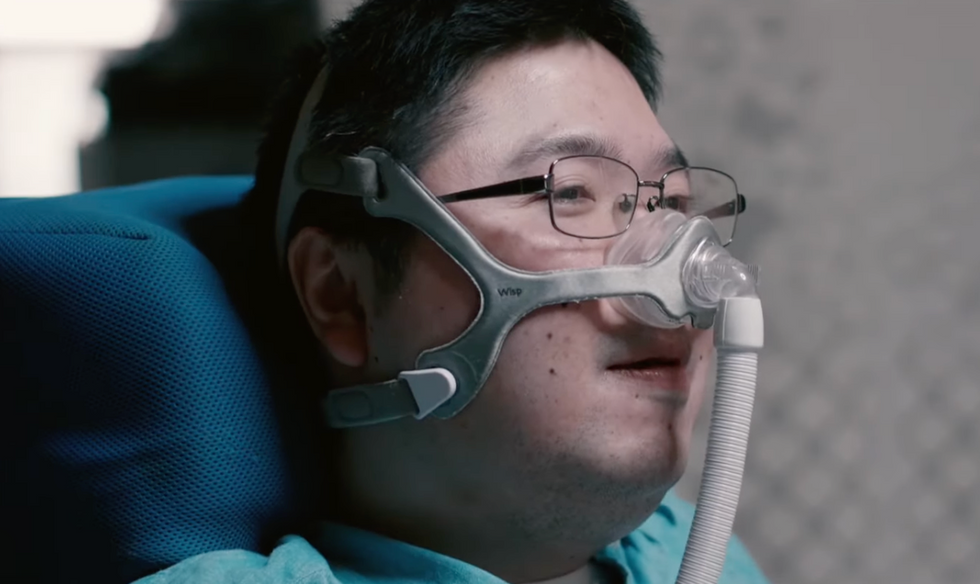
Shota, Kuwahara, a DAWN employee with muscular dystrophy, agrees. "There are many difficulties in my daily life, but I believe my life has a purpose and is not being wasted," he says. "Being useful, able to help other people, even feeling needed by others, is so motivational."
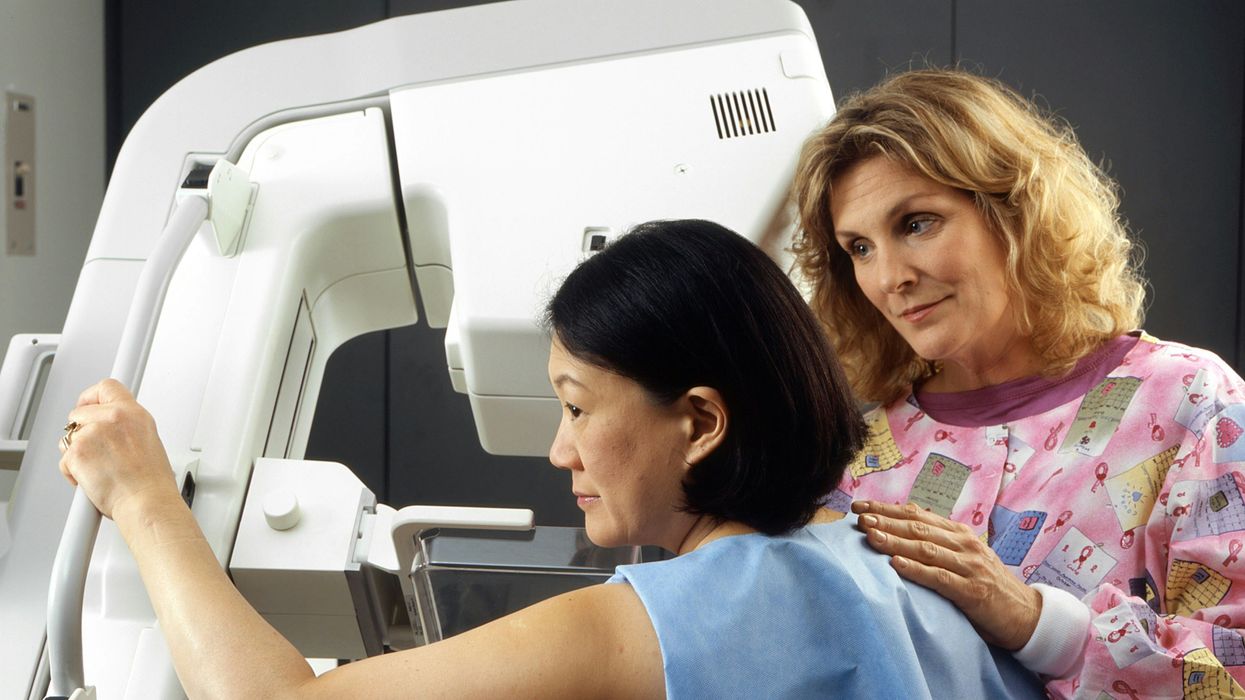
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.