Anyone with a Computer Can Join the Fight Against COVID-19 Right Now

A participatory research project has evolved in the world's most powerful networked supercomputer to identify drug targets for COVID-19.
With millions of people left feeling helpless as COVID-19 sweeps across the U.S. and the rest of the planet, there is one way in which absolutely anyone can help fight the pandemic -- all you need is a computer and an Internet connection.
"The more donors that participate, the more science we're able to do."
The Folding@home project allows members of the public to contribute a portion of their computing power to a gigantic virtual network which has mushroomed over the past month to become the most powerful supercomputer on the planet.
As of April 6, more than one million people across the globe have donated some of their home computing resources to the project. Combined, this gives Folding@home processing powers that dwarf even NASA and IBM's most powerful devices. To join, all you have to do is go to this website and click 'Download Now' to load the Folding@home software on your computer. This runs in the background, and only adds your unused computing power to the project, so it will not drain resources from tasks you're trying to do.
"It's totally crazy," said Vincent Voelz, associate professor of chemistry at Temple University, Philadelphia, and one of the scientists leading the project. "A month ago, we had around 30,000 to 40,000 participants. And then last week, it rose up 400,000 and now we've hit a million. But the more donors that participate, the more science we're able to do."
Voelz and the other scientists behind Folding@home are using these vast resources to model the ever-changing shapes of the coronavirus's proteins, in the hopes of identifying vulnerabilities or 'pockets' in its structure that can be targeted with new drugs.
One of the reasons it's difficult to find treatments for viruses like COVID-19 and Ebola is because the proteins, the innate building blocks of the viral structure, have notoriously smooth surfaces, making it hard for drugs to bind to them.
But viral proteins don't stay still. They are constantly evolving and changing shape as the atoms within push and pull against each other. Having a supercomputer enables scientists to simulate all these different shapes, revealing potential weaknesses which were not immediately visible. And the more powerful the supercomputer, the faster these simulations can happen.
"Simulating these protein motions also enables us to answer basic questions such as what makes this new coronavirus strain different from previous strains," said Voelz. "Is there something about the dynamics of these proteins that makes it more virulent?"
Finding a genuinely novel drug for COVID-19 is particularly critical.
Once they have identified suitable pockets within the proteins of COVID-19, the Folding@home scientists can then take the many compounds being identified by chemists around the world as potential drugs, and try to predict which ones will stand the best chance of binding to those pockets and inhibiting the virus's ability to invade and take over human cells.
"We have so much bandwidth now with Folding@home that we really think we can make a dent with screening these, and prioritizing which compounds are then going to get experimentally tested," said Voeltz.
The team are particularly hopeful they can succeed, having already used the supercomputer to identify a new vulnerability in the Ebola virus, which could go on to yield a new treatment for the disease.
Finding a genuinely novel drug for COVID-19 is particularly critical. While researchers are also looking at repurposing existing medications, like the antimalarials Hydroxychloroquine and Chloroquine (which have just been approved by the FDA for emergency use in coronavirus patients), concerns remain about the safety of these treatments. Researchers at the Mayo Clinic recently warned that the use of these drugs could have the side effect of inducing heart problems and run the risk of sudden cardiac arrest.
But with the death toll increasing by the day, speed is of the essence. Voelz explains that the scientific community has been left playing catch-up, because a drug was never actually developed for the original SARS outbreak in the early 2000s. The enormous computational power of the Folding@home project has the potential to allow scientists to quickly answer some of the key questions needed to get a new treatment into the pipeline.
"We don't have a SARS drug for whatever reason," said Voelz. "So the missing ingredient really, is the basic science to reveal possible drug targets and then the pharma can take that information and do the engineering work and optimizing and clinically testing drugs. But we now have a lot of basic science going on in response to this pandemic."
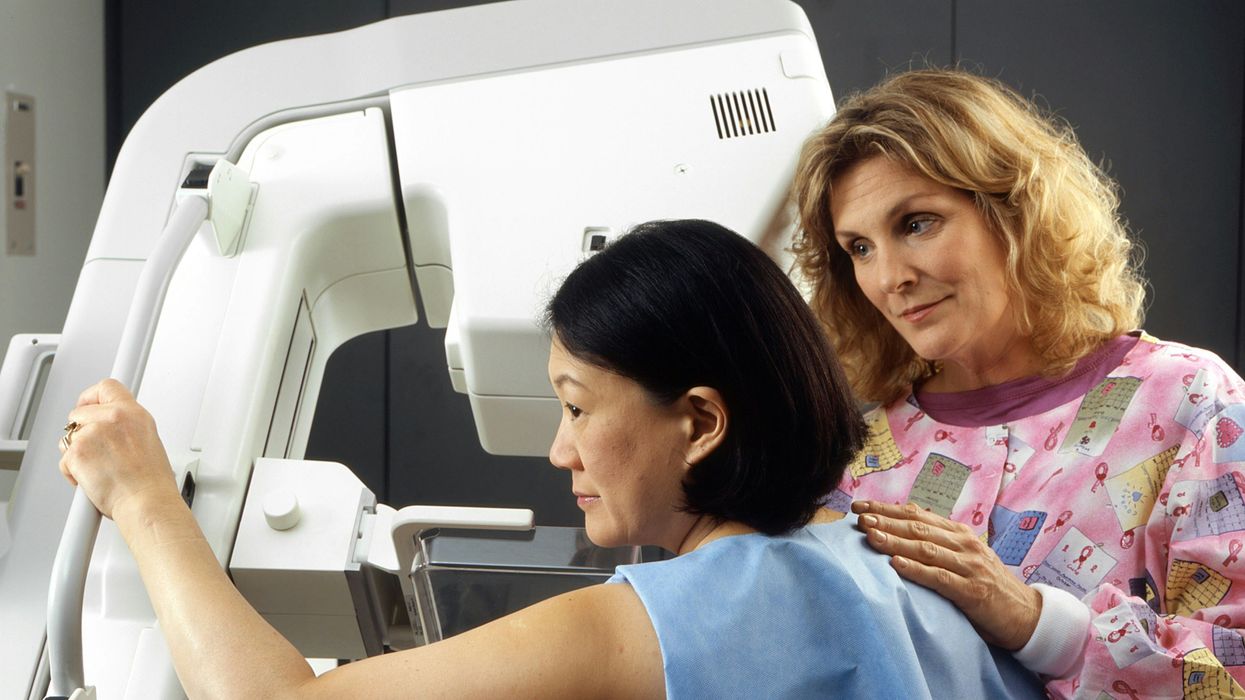
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.
Urinary tract infections account for more than 8 million trips to the doctor each year.
Few things are more painful than a urinary tract infection (UTI). Common in men and women, these infections account for more than 8 million trips to the doctor each year and can cause an array of uncomfortable symptoms, from a burning feeling during urination to fever, vomiting, and chills. For an unlucky few, UTIs can be chronic—meaning that, despite treatment, they just keep coming back.
But new research, presented at the European Association of Urology (EAU) Congress in Paris this week, brings some hope to people who suffer from UTIs.
Clinicians from the Royal Berkshire Hospital presented the results of a long-term, nine-year clinical trial where 89 men and women who suffered from recurrent UTIs were given an oral vaccine called MV140, designed to prevent the infections. Every day for three months, the participants were given two sprays of the vaccine (flavored to taste like pineapple) and then followed over the course of nine years. Clinicians analyzed medical records and asked the study participants about symptoms to check whether any experienced UTIs or had any adverse reactions from taking the vaccine.
The results showed that across nine years, 48 of the participants (about 54%) remained completely infection-free. On average, the study participants remained infection free for 54.7 months—four and a half years.
“While we need to be pragmatic, this vaccine is a potential breakthrough in preventing UTIs and could offer a safe and effective alternative to conventional treatments,” said Gernot Bonita, Professor of Urology at the Alta Bro Medical Centre for Urology in Switzerland, who is also the EAU Chairman of Guidelines on Urological Infections.
The news comes as a relief not only for people who suffer chronic UTIs, but also to doctors who have seen an uptick in antibiotic-resistant UTIs in the past several years. Because UTIs usually require antibiotics, patients run the risk of developing a resistance to the antibiotics, making infections more difficult to treat. A preventative vaccine could mean less infections, less antibiotics, and less drug resistance overall.
“Many of our participants told us that having the vaccine restored their quality of life,” said Dr. Bob Yang, Consultant Urologist at the Royal Berkshire NHS Foundation Trust, who helped lead the research. “While we’re yet to look at the effect of this vaccine in different patient groups, this follow-up data suggests it could be a game-changer for UTI prevention if it’s offered widely, reducing the need for antibiotic treatments.”