The Age of DNA-Based Dating Is Here

A woman gives a DNA sample at a Pheramor-sponsored event.
Brittany Barreto first got the idea to make a DNA-based dating platform nearly 10 years ago when she was in a college seminar on genetics. She joked that it would be called GeneHarmony.com.
Pheramor and startups, like DNA Romance and Instant Chemistry, both based in Canada, claim to match you to a romantic partner based on your genetics.
The idea stuck with her while she was getting her PhD in genetics at Baylor College of Medicine, and in March 2018, she launched Pheramor, a dating app that measures compatibility based on physical chemistry and what the company calls "social alignment."
"I wanted to use genetics and science to help people connect more. Our world is so hungry for connection," says Barreto, who serves as Pheramor's CEO.
With the direct-to-consumer genetic testing market booming, more and more companies are looking to capitalize on the promise of DNA-based services. Pheramor and startups, like DNA Romance and Instant Chemistry, both based in Canada, claim to match you to a romantic partner based on your genetics. It's an intriguing alternative to swiping left or right in hopes of finding someone you're not only physically attracted to but actually want to date. Experts say the science behind such apps isn't settled though.
For $40, Pheramor sends you a DNA kit to swab the inside of your cheek. After you mail in your sample, Pheramor analyzes your saliva for 11 different HLA genes, a fraction of the more than 200 genes that are thought to make up the human HLA complex. These genes make proteins that regulate the immune system by helping protect against invading pathogens.
It takes three to four weeks to get the results backs. In the meantime, users can still download the app and start using it before their DNA results are ready. The app asks users to link their social media accounts, which are fed into an algorithm that calculates a "social alignment." The algorithm takes into account the hashtags you use, your likes, check-ins, posts, and accounts you follow on Facebook, Twitter, and Instagram.
The DNA test results and social alignment algorithm are used to calculate a compatibility percentage between zero and 100. Barreto said she couldn't comment on how much of that score is influenced by the algorithm and how much comes from what the company calls genetic attraction. "DNA is not destiny," she says. "It's not like you're going to swab and I'll send you your soulmate."
Despite its name, Pheramor doesn't actually measure pheromones, chemicals released by animals that affect the behavior of others of the same species. That's because human pheromones have yet to be identified, though they've been discovered throughout the animal kingdom in moths, mice, rabbits, pigs, and many other insects and mammals. The HLA genes Pheramor analyzes instead are the human version of the major histocompatibility complex (MHC), a gene group found in many species.
The connection between HLA type and attraction goes back to the 1970s, when researchers found that inbred male mice preferred to mate with female mice with a different MHC rather than inbred female mice with similar immune system genes. The researchers concluded that this mating preference was linked to smell. The idea is that choosing a mate with different MHC genes gives animals an evolutionary advantage in terms of immune system defense.
The couples who had more dissimilar HLA types reported a more satisfied sex life and satisfied partnership, but it was a small effect.
In the 1990s, Swiss scientists wanted to see if body odor also had an effect on human attraction. In a famous experiment known as the "sweaty T-shirt study", they recruited 49 women to sniff sweaty, unwashed T-shirts from 44 men and put each in a box with a smelling hole and describe the odors of every shirt. The study found that women preferred the scents of T-shirts worn by men who were immunologically different from them compared to men whose HLA genes were similar to their own.
"The idea is, if you are very similar with your partner in HLA type then your offspring is similar in terms of HLA. This reduces your resistance against pathogens," says Illona Croy, a psychologist at the Technical University of Dresden who has studied HLA type in relation to sexual attraction in humans.
In a 2016 study Pheramor cites on its website, Croy and her colleagues tested the HLA types of 250 couples—all of them university students—and asked them how satisfied they were with their partnerships, with their sex lives, and with the odors of their partners. The couples who had more dissimilar HLA types reported a more satisfied sex life and satisfied partnership, but Croy cautions that it was a small effect. "It's not like they were super satisfied or not satisfied at all. It's a slight difference," she says.
Croy says we're much more likely to choose a partner based on appearance, sense of humor, intelligence and common interests.
Other studies have reported no preference for HLA difference in sexual attraction. Tristram Wyatt, a zoologist at the University of Oxford in the U.K. who studies animal pheromones, says it's been difficult to replicate the original T-shirt study. And one of the caveats of the original study is that women who were taking birth control pills preferred men who were more immunologically similar.
"Certainly, we learn to really like the smell of our partners," Wyatt says. "Whether it's the reason for choosing them in the first place, we really don't know."
Wyatt says he's skeptical of DNA-based dating apps because there are many subtypes of HLA genes, meaning there's a fairly low chance that your HLA type and your romantic partner's would be an exact match, anyway. It's why finding a suitable match for a bone marrow transplant is difficult; a donor's HLA type has to be the same as the recipient's.
"What it means is that since we're all different, it's hard statistically to say who the best match will be," he says.
DNA-based dating apps haven't yet gone mainstream, but some people seem willing to give them a try. Since Pheramor's launch a little over a year ago, about 10,000 people have signed up to use the app, about half of which have taken the DNA test, Barreto says. By comparison, an estimated 50 million people use Tinder, which has been around since 2012, and about 40 million people are on Bumble, which was released in 2014.
In April, Barreto launched a second service, this one for couples, called WeHaveChemistry.com. A $139 kit includes two genetic tests, one for you and your partner, and a detailed DNA report on your sexual compatibility.
Unlike the Phermor app, WeHaveChemistry doesn't provide users with a numeric combability score but instead makes personalized recommendations based on your genetic results. For instance, if the DNA test shows that your HLA genes are similar, Barreto says, "We might recommend pheromone colognes, working out together, or not showering before bed to get your juices running."
Despite her own research on HLA and sexual compatibility, Croy isn't sure how knowing HLA type will help couples. However, some researchers are doing studies on whether HLA types are related to certain cases of infertility, and this is where a genetic test might be very useful, says Croy.
"Otherwise, I think it doesn't matter whether we're HLA compatible or not," she says. "It might give you one possible explanation about why your sexual life isn't as satisfactory as it could be, but there are many other factors that play a role."
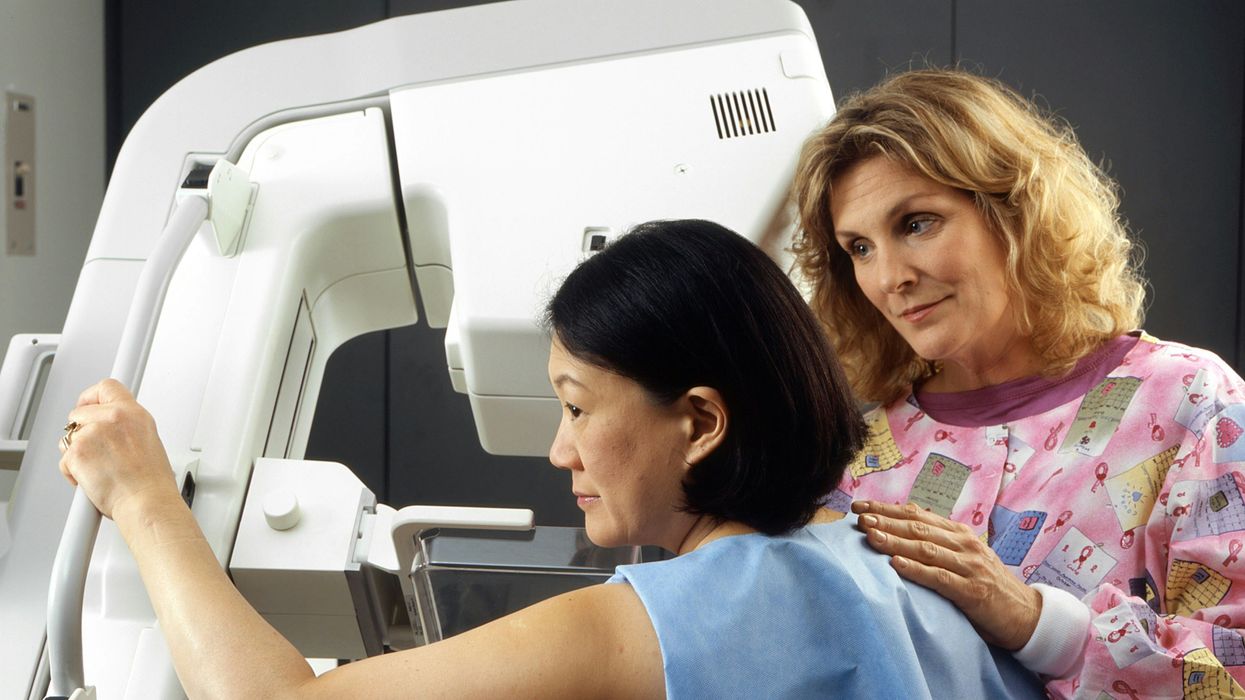
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.
Urinary tract infections account for more than 8 million trips to the doctor each year.
Few things are more painful than a urinary tract infection (UTI). Common in men and women, these infections account for more than 8 million trips to the doctor each year and can cause an array of uncomfortable symptoms, from a burning feeling during urination to fever, vomiting, and chills. For an unlucky few, UTIs can be chronic—meaning that, despite treatment, they just keep coming back.
But new research, presented at the European Association of Urology (EAU) Congress in Paris this week, brings some hope to people who suffer from UTIs.
Clinicians from the Royal Berkshire Hospital presented the results of a long-term, nine-year clinical trial where 89 men and women who suffered from recurrent UTIs were given an oral vaccine called MV140, designed to prevent the infections. Every day for three months, the participants were given two sprays of the vaccine (flavored to taste like pineapple) and then followed over the course of nine years. Clinicians analyzed medical records and asked the study participants about symptoms to check whether any experienced UTIs or had any adverse reactions from taking the vaccine.
The results showed that across nine years, 48 of the participants (about 54%) remained completely infection-free. On average, the study participants remained infection free for 54.7 months—four and a half years.
“While we need to be pragmatic, this vaccine is a potential breakthrough in preventing UTIs and could offer a safe and effective alternative to conventional treatments,” said Gernot Bonita, Professor of Urology at the Alta Bro Medical Centre for Urology in Switzerland, who is also the EAU Chairman of Guidelines on Urological Infections.
The news comes as a relief not only for people who suffer chronic UTIs, but also to doctors who have seen an uptick in antibiotic-resistant UTIs in the past several years. Because UTIs usually require antibiotics, patients run the risk of developing a resistance to the antibiotics, making infections more difficult to treat. A preventative vaccine could mean less infections, less antibiotics, and less drug resistance overall.
“Many of our participants told us that having the vaccine restored their quality of life,” said Dr. Bob Yang, Consultant Urologist at the Royal Berkshire NHS Foundation Trust, who helped lead the research. “While we’re yet to look at the effect of this vaccine in different patient groups, this follow-up data suggests it could be a game-changer for UTI prevention if it’s offered widely, reducing the need for antibiotic treatments.”