Meet the Scientists on the Frontlines of Protecting Humanity from a Man-Made Pathogen

From left: Jean Peccoud, Randall Murch, and Neeraj Rao.
Jean Peccoud wasn't expecting an email from the FBI. He definitely wasn't expecting the agency to invite him to a meeting. "My reaction was, 'What did I do wrong to be on the FBI watch list?'" he recalls.
You use those blueprints for white-hat research—which is, indeed, why the open blueprints exist—or you can do the same for a black-hat attack.
He didn't know what the feds could possibly want from him. "I was mostly scared at this point," he says. "I was deeply disturbed by the whole thing."
But he decided to go anyway, and when he traveled to San Francisco for the 2008 gathering, the reason for the e-vite became clear: The FBI was reaching out to researchers like him—scientists interested in synthetic biology—in anticipation of the potential nefarious uses of this technology. "The whole purpose of the meeting was, 'Let's start talking to each other before we actually need to talk to each other,'" says Peccoud, now a professor of chemical and biological engineering at Colorado State University. "'And let's make sure next time you get an email from the FBI, you don't freak out."
Synthetic biology—which Peccoud defines as "the application of engineering methods to biological systems"—holds great power, and with that (as always) comes great responsibility. When you can synthesize genetic material in a lab, you can create new ways of diagnosing and treating people, and even new food ingredients. But you can also "print" the genetic sequence of a virus or virulent bacterium.
And while it's not easy, it's also not as hard as it could be, in part because dangerous sequences have publicly available blueprints. You use those blueprints for white-hat research—which is, indeed, why the open blueprints exist—or you can do the same for a black-hat attack. You could synthesize a dangerous pathogen's code on purpose, or you could unwittingly do so because someone tampered with your digital instructions. Ordering synthetic genes for viral sequences, says Peccoud, would likely be more difficult today than it was a decade ago.
"There is more awareness of the industry, and they are taking this more seriously," he says. "There is no specific regulation, though."
Trying to lock down the interconnected machines that enable synthetic biology, secure its lab processes, and keep dangerous pathogens out of the hands of bad actors is part of a relatively new field: cyberbiosecurity, whose name Peccoud and colleagues introduced in a 2018 paper.
Biological threats feel especially acute right now, during the ongoing pandemic. COVID-19 is a natural pathogen -- not one engineered in a lab. But future outbreaks could start from a bug nature didn't build, if the wrong people get ahold of the right genetic sequences, and put them in the right sequence. Securing the equipment and processes that make synthetic biology possible -- so that doesn't happen -- is part of why the field of cyberbiosecurity was born.
The Origin Story
It is perhaps no coincidence that the FBI pinged Peccoud when it did: soon after a journalist ordered a sequence of smallpox DNA and wrote, for The Guardian, about how easy it was. "That was not good press for anybody," says Peccoud. Previously, in 2002, the Pentagon had funded SUNY Stonybrook researchers to try something similar: They ordered bits of polio DNA piecemeal and, over the course of three years, strung them together.
Although many years have passed since those early gotchas, the current patchwork of regulations still wouldn't necessarily prevent someone from pulling similar tricks now, and the technological systems that synthetic biology runs on are more intertwined — and so perhaps more hackable — than ever. Researchers like Peccoud are working to bring awareness to those potential problems, to promote accountability, and to provide early-detection tools that would catch the whiff of a rotten act before it became one.
Peccoud notes that if someone wants to get access to a specific pathogen, it is probably easier to collect it from the environment or take it from a biodefense lab than to whip it up synthetically. "However, people could use genetic databases to design a system that combines different genes in a way that would make them dangerous together without each of the components being dangerous on its own," he says. "This would be much more difficult to detect."
After his meeting with the FBI, Peccoud grew more interested in these sorts of security questions. So he was paying attention when, in 2010, the Department of Health and Human Services — now helping manage the response to COVID-19 — created guidance for how to screen synthetic biology orders, to make sure suppliers didn't accidentally send bad actors the sequences that make up bad genomes.
Guidance is nice, Peccoud thought, but it's just words. He wanted to turn those words into action: into a computer program. "I didn't know if it was something you can run on a desktop or if you need a supercomputer to run it," he says. So, one summer, he tasked a team of student researchers with poring over the sentences and turning them into scripts. "I let the FBI know," he says, having both learned his lesson and wanting to get in on the game.
Peccoud later joined forces with Randall Murch, a former FBI agent and current Virginia Tech professor, and a team of colleagues from both Virginia Tech and the University of Nebraska-Lincoln, on a prototype project for the Department of Defense. They went into a lab at the University of Nebraska at Lincoln and assessed all its cyberbio-vulnerabilities. The lab develops and produces prototype vaccines, therapeutics, and prophylactic components — exactly the kind of place that you always, and especially right now, want to keep secure.
"We were creating wiki of all these nasty things."
The team found dozens of Achilles' heels, and put them in a private report. Not long after that project, the two and their colleagues wrote the paper that first used the term "cyberbiosecurity." A second paper, led by Murch, came out five months later and provided a proposed definition and more comprehensive perspective on cyberbiosecurity. But although it's now a buzzword, it's the definition, not the jargon, that matters. "Frankly, I don't really care if they call it cyberbiosecurity," says Murch. Call it what you want: Just pay attention to its tenets.
A Database of Scary Sequences
Peccoud and Murch, of course, aren't the only ones working to screen sequences and secure devices. At the nonprofit Battelle Memorial Institute in Columbus, Ohio, for instance, scientists are working on solutions that balance the openness inherent to science and the closure that can stop bad stuff. "There's a challenge there that you want to enable research but you want to make sure that what people are ordering is safe," says the organization's Neeraj Rao.
Rao can't talk about the work Battelle does for the spy agency IARPA, the Intelligence Advanced Research Projects Activity, on a project called Fun GCAT, which aims to use computational tools to deep-screen gene-sequence orders to see if they pose a threat. It can, though, talk about a twin-type internal project: ThreatSEQ (pronounced, of course, "threat seek").
The project started when "a government customer" (as usual, no one will say which) asked Battelle to curate a list of dangerous toxins and pathogens, and their genetic sequences. The researchers even started tagging sequences according to their function — like whether a particular sequence is involved in a germ's virulence or toxicity. That helps if someone is trying to use synthetic biology not to gin up a yawn-inducing old bug but to engineer a totally new one. "How do you essentially predict what the function of a novel sequence is?" says Rao. You look at what other, similar bits of code do.
"We were creating wiki of all these nasty things," says Rao. As they were working, they realized that DNA manufacturers could potentially scan in sequences that people ordered, run them against the database, and see if anything scary matched up. Kind of like that plagiarism software your college professors used.
Battelle began offering their screening capability, as ThreatSEQ. When customers -- like, currently, Twist Bioscience -- throw their sequences in, and get a report back, the manufacturers make the final decision about whether to fulfill a flagged order — whether, in the analogy, to give an F for plagiarism. After all, legitimate researchers do legitimately need to have DNA from legitimately bad organisms.
"Maybe it's the CDC," says Rao. "If things check out, oftentimes [the manufacturers] will fulfill the order." If it's your aggrieved uncle seeking the virulent pathogen, maybe not. But ultimately, no one is stopping the manufacturers from doing so.
Beyond that kind of tampering, though, cyberbiosecurity also includes keeping a lockdown on the machines that make the genetic sequences. "Somebody now doesn't need physical access to infrastructure to tamper with it," says Rao. So it needs the same cyber protections as other internet-connected devices.
Scientists are also now using DNA to store data — encoding information in its bases, rather than into a hard drive. To download the data, you sequence the DNA and read it back into a computer. But if you think like a bad guy, you'd realize that a bad guy could then, for instance, insert a computer virus into the genetic code, and when the researcher went to nab her data, her desktop would crash or infect the others on the network.
Something like that actually happened in 2017 at the USENIX security symposium, an annual programming conference: Researchers from the University of Washington encoded malware into DNA, and when the gene sequencer assembled the DNA, it corrupted the sequencer's software, then the computer that controlled it.
"This vulnerability could be just the opening an adversary needs to compromise an organization's systems," Inspirion Biosciences' J. Craig Reed and Nicolas Dunaway wrote in a paper for Frontiers in Bioengineering and Biotechnology, included in an e-book that Murch edited called Mapping the Cyberbiosecurity Enterprise.
Where We Go From Here
So what to do about all this? That's hard to say, in part because we don't know how big a current problem any of it poses. As noted in Mapping the Cyberbiosecurity Enterprise, "Information about private sector infrastructure vulnerabilities or data breaches is protected from public release by the Protected Critical Infrastructure Information (PCII) Program," if the privateers share the information with the government. "Government sector vulnerabilities or data breaches," meanwhile, "are rarely shared with the public."
"What I think is encouraging right now is the fact that we're even having this discussion."
The regulations that could rein in problems aren't as robust as many would like them to be, and much good behavior is technically voluntary — although guidelines and best practices do exist from organizations like the International Gene Synthesis Consortium and the National Institute of Standards and Technology.
Rao thinks it would be smart if grant-giving agencies like the National Institutes of Health and the National Science Foundation required any scientists who took their money to work with manufacturing companies that screen sequences. But he also still thinks we're on our way to being ahead of the curve, in terms of preventing print-your-own bioproblems: "What I think is encouraging right now is the fact that we're even having this discussion," says Rao.
Peccoud, for his part, has worked to keep such conversations going, including by doing training for the FBI and planning a workshop for students in which they imagine and work to guard against the malicious use of their research. But actually, Peccoud believes that human error, flawed lab processes, and mislabeled samples might be bigger threats than the outside ones. "Way too often, I think that people think of security as, 'Oh, there is a bad guy going after me,' and the main thing you should be worried about is yourself and errors," he says.
Murch thinks we're only at the beginning of understanding where our weak points are, and how many times they've been bruised. Decreasing those contusions, though, won't just take more secure systems. "The answer won't be technical only," he says. It'll be social, political, policy-related, and economic — a cultural revolution all its own.
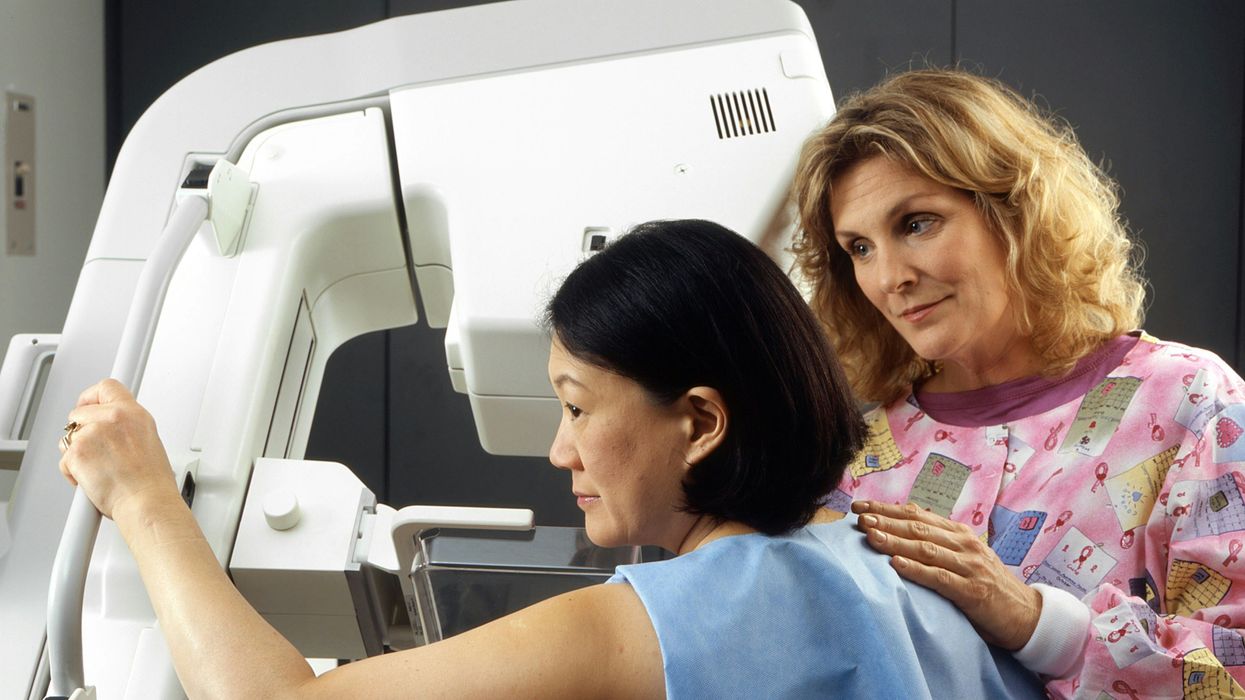
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.
Urinary tract infections account for more than 8 million trips to the doctor each year.
Few things are more painful than a urinary tract infection (UTI). Common in men and women, these infections account for more than 8 million trips to the doctor each year and can cause an array of uncomfortable symptoms, from a burning feeling during urination to fever, vomiting, and chills. For an unlucky few, UTIs can be chronic—meaning that, despite treatment, they just keep coming back.
But new research, presented at the European Association of Urology (EAU) Congress in Paris this week, brings some hope to people who suffer from UTIs.
Clinicians from the Royal Berkshire Hospital presented the results of a long-term, nine-year clinical trial where 89 men and women who suffered from recurrent UTIs were given an oral vaccine called MV140, designed to prevent the infections. Every day for three months, the participants were given two sprays of the vaccine (flavored to taste like pineapple) and then followed over the course of nine years. Clinicians analyzed medical records and asked the study participants about symptoms to check whether any experienced UTIs or had any adverse reactions from taking the vaccine.
The results showed that across nine years, 48 of the participants (about 54%) remained completely infection-free. On average, the study participants remained infection free for 54.7 months—four and a half years.
“While we need to be pragmatic, this vaccine is a potential breakthrough in preventing UTIs and could offer a safe and effective alternative to conventional treatments,” said Gernot Bonita, Professor of Urology at the Alta Bro Medical Centre for Urology in Switzerland, who is also the EAU Chairman of Guidelines on Urological Infections.
The news comes as a relief not only for people who suffer chronic UTIs, but also to doctors who have seen an uptick in antibiotic-resistant UTIs in the past several years. Because UTIs usually require antibiotics, patients run the risk of developing a resistance to the antibiotics, making infections more difficult to treat. A preventative vaccine could mean less infections, less antibiotics, and less drug resistance overall.
“Many of our participants told us that having the vaccine restored their quality of life,” said Dr. Bob Yang, Consultant Urologist at the Royal Berkshire NHS Foundation Trust, who helped lead the research. “While we’re yet to look at the effect of this vaccine in different patient groups, this follow-up data suggests it could be a game-changer for UTI prevention if it’s offered widely, reducing the need for antibiotic treatments.”