Genome Reading and Editing Tools for All

An open book representing the ability to read the human genome.
In 2006, the cover of Scientific American was "Know Your DNA" and the inside story was "Genomes for All." Today, we are closer to that goal than ever. Making it affordable for everyone to understand and change their DNA will fundamentally alter how we manage diseases, how we conduct clinical research, and even how we select a mate.
A frequent line of questions on the topic of making genome reading affordable is: Do we need to read the whole genome in order to accurately predict disease risk?
Since 2006, we have driven the cost of reading a human genome down from $3 billion to $600. To aid interpretation and research to produce new diagnostics and therapeutics, my research team at Harvard initiated the Personal Genome Project and later, Openhumans.org. This has demonstrated international informed consent for human genomes, and diverse environmental and trait data can be distributed freely. This is done with no strings attached in a manner analogous to Wikipedia. Cell lines from that project are similarly freely available for experiments on synthetic biology, gene therapy and human developmental biology. DNA from those cells have been chosen by the US National Institute of Standards and Technology and the Food and Drug Administration to be the key federal standards for the human genome.
A frequent line of questions on the topic of making genome reading affordable is: Do we need to read the whole genome in order to accurately predict disease risk? Can we just do most commonly varying parts of the genome, which constitute only a tiny fraction of a percent? Or just the most important parts encoding the proteins or 'exome,' which constitute about one percent of the genome? The commonly varying parts of the genome are poor predictors of serious genetic diseases and the exomes don't detect DNA rearrangements which often wipe out gene function when they occur in non-coding regions within genes. Since the cost of the exome is not one percent of the whole genome cost, but nearly identical ($600), missing an impactful category of mutants is really not worth it. So the answer is yes, we should read the whole genome to glean comprehensively meaningful information.
In parallel to the reading revolution, we have dropped the price of DNA synthesis by a similar million-fold and made genome editing tools close to free.
WRITING
In parallel to the reading revolution, we have dropped the price of DNA synthesis by a similar million-fold and made genome editing tools like CRISPR, TALE and MAGE close to free by distributing them through the non-profit Addgene.org. Gene therapies are already curing blindness in children and cancer in adults, and hopefully soon infectious diseases and hemoglobin diseases like sickle cell anemia. Nevertheless, gene therapies are (so far) the most expensive class of drugs in history (about $1 million dollars per dose).
This is in large part because the costs of proving safety and efficacy in a randomized clinical trial are high and that cost is spread out only over the people that benefit (aka the denominator). Striking growth is evident in such expensive hyper-personalized therapies ever since the "Orphan Drug Act of 1983." For the most common disease, aging (which kills 90 percent of people in wealthy regions of the world), the denominator is maximal and the cost of the drugs should be low as genetic interventions to combat aging become available in the next ten years. But what can we do about rarer diseases with cheap access to genome reading and editing tools? Try to prevent them in the first place.
A huge fraction of these births is preventable if unaffected carriers of such diseases do not mate.
ARITHMETIC
While the cost of reading has plummeted, the value of knowing your genome is higher than ever. About 5 percent of births result in extreme medical trauma over a person's lifetime due to rare genetic diseases. Even without gene therapy, these cost the family and society more than a million dollars in drugs, diagnostics and instruments, extra general care, loss of income for the affected individual and other family members, plus pain and anxiety of the "medical odyssey" often via dozens of mystified physicians. A huge fraction of these births is preventable if unaffected carriers of such diseases do not mate.
The non-profit genetic screening organization, Dor Yeshorim (established in 1983), has shown that this is feasible by testing for Tay–Sachs disease, Familial dysautonomia, Cystic fibrosis, Canavan disease, Glycogen storage disease (type 1), Fanconi anemia (type C), Bloom syndrome, Niemann–Pick disease, Mucolipidosis type IV. This is often done at the pre-marital, matchmaking phase, which can reduce the frequency of natural or induced abortions. Such matchmaking can be done in such a way that no one knows the carrier status of any individual in the system. In addition to those nine tests, many additional diseases can be picked up by whole genome sequencing. No person can know in advance that they are exemptfrom these risks.
Furthermore, concerns about rare "false positives" is far less at the stage of matchmaking than at the stage of prenatal testing, since the latter could involve termination of a healthy fetus, while the former just means that you restrict your dating to 90 percent of the population. In order to scale this up from 13 million Ashkenazim and Sephardim to billions in diverse cultures, we will likely see new computer security, encryption, blockchain and matchmaking tools.
Once the diseases are eradicated from our population, the interventions can be said to impact not only the current population, but all subsequent generations.
THE FUTURE
As reading and writing become exponentially more affordable and reliable, we can tackle equitable distribution, but there remain issues of education and security. Society, broadly (insurers, health care providers, governments) should be able to see a roughly 12-fold return on their investment of $1800 per person ($600 each for raw data, interpretation and incentivizing the participant) by saving $1 million per diseased child per 20 families. Everyone will have free access to their genome information and software to guide their choices in precision medicines, mates and participation in biomedical research studies.
In terms of writing and editing, if delivery efficiency and accuracy keep improving, then pill or aerosol formulations of gene therapies -- even non-prescription, veterinary or home-made versions -- are not inconceivable. Preventions tends to be more affordable and more humane than cures. If gene therapies provide prevention of diseases of aging, cancer and cognitive decline, they might be considered "enhancement," but not necessarily more remarkable than past preventative strategies, like vaccines against HPV-cancer, smallpox and polio. Whether we're overcoming an internal genetic flaw or an external infectious disease, the purpose is the same: to minimize human suffering. Once the diseases are eradicated from our population, the interventions can be said to impact not only the current population, but all subsequent generations. This reminds us that we need to listen carefully, educate each other and proactively imagine and deflect likely, and even unlikely, unintended consequences, including stigmatization of the last few unprotected individuals.
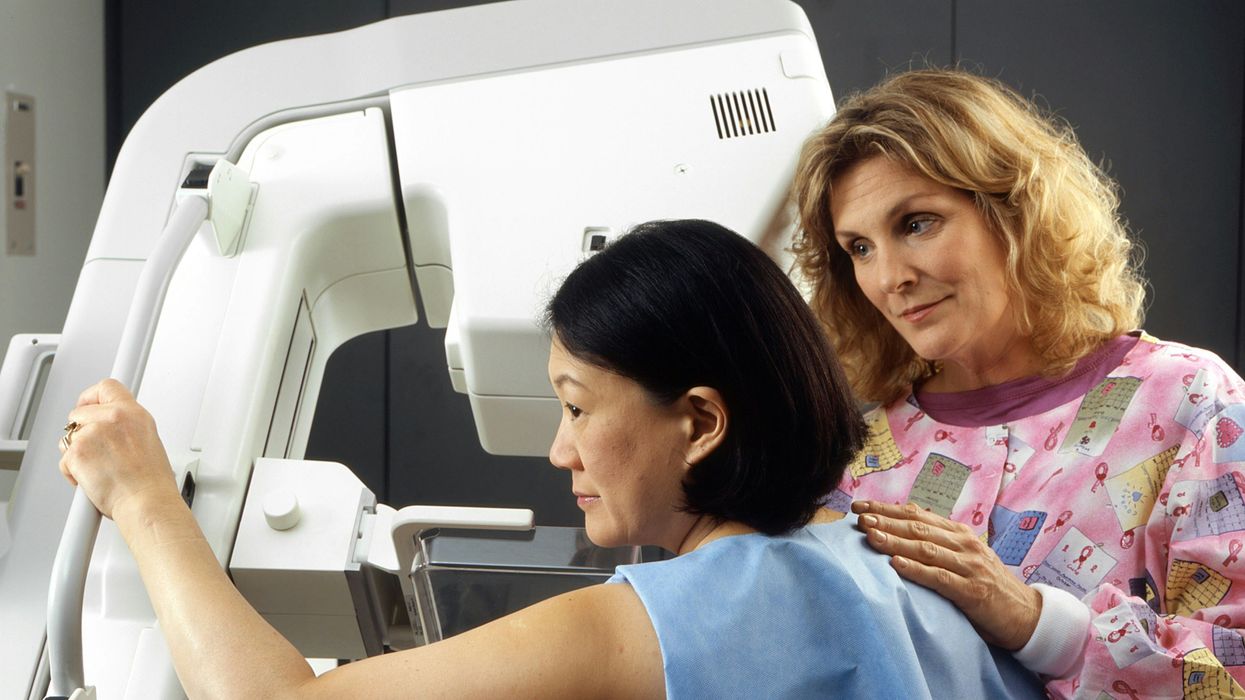
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.
Urinary tract infections account for more than 8 million trips to the doctor each year.
Few things are more painful than a urinary tract infection (UTI). Common in men and women, these infections account for more than 8 million trips to the doctor each year and can cause an array of uncomfortable symptoms, from a burning feeling during urination to fever, vomiting, and chills. For an unlucky few, UTIs can be chronic—meaning that, despite treatment, they just keep coming back.
But new research, presented at the European Association of Urology (EAU) Congress in Paris this week, brings some hope to people who suffer from UTIs.
Clinicians from the Royal Berkshire Hospital presented the results of a long-term, nine-year clinical trial where 89 men and women who suffered from recurrent UTIs were given an oral vaccine called MV140, designed to prevent the infections. Every day for three months, the participants were given two sprays of the vaccine (flavored to taste like pineapple) and then followed over the course of nine years. Clinicians analyzed medical records and asked the study participants about symptoms to check whether any experienced UTIs or had any adverse reactions from taking the vaccine.
The results showed that across nine years, 48 of the participants (about 54%) remained completely infection-free. On average, the study participants remained infection free for 54.7 months—four and a half years.
“While we need to be pragmatic, this vaccine is a potential breakthrough in preventing UTIs and could offer a safe and effective alternative to conventional treatments,” said Gernot Bonita, Professor of Urology at the Alta Bro Medical Centre for Urology in Switzerland, who is also the EAU Chairman of Guidelines on Urological Infections.
The news comes as a relief not only for people who suffer chronic UTIs, but also to doctors who have seen an uptick in antibiotic-resistant UTIs in the past several years. Because UTIs usually require antibiotics, patients run the risk of developing a resistance to the antibiotics, making infections more difficult to treat. A preventative vaccine could mean less infections, less antibiotics, and less drug resistance overall.
“Many of our participants told us that having the vaccine restored their quality of life,” said Dr. Bob Yang, Consultant Urologist at the Royal Berkshire NHS Foundation Trust, who helped lead the research. “While we’re yet to look at the effect of this vaccine in different patient groups, this follow-up data suggests it could be a game-changer for UTI prevention if it’s offered widely, reducing the need for antibiotic treatments.”