Scientists aim to preserve donkeys, one frozen embryo at a time
Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.

In Ethiopia, Samuna’s three donkeys help her transport produce to market and to collect the water essential to her family, neighbours and livestock. Donkeys are more endangered than people realize, experts say.
Every day for a week in 2022, Andres Gambini, a veterinarian and senior lecturer in animal science at the University of Queensland in Australia, walked into his lab—and headed straight to the video camera. Trained on an array of about 50 donkey embryos, all created by Gambini’s manual in vitro fertilization, or IVF, the camera kept an eye on their developmental progress. To eventually create a viable embryo that could be implanted into a female donkey, the embryos’ cells had to keep dividing, first in two, then in four and so on.
But the embryos weren’t cooperating. Some would start splitting up only to stop a day or two later, and others wouldn’t start at all. Every day he came in, Gambini saw fewer and fewer dividing embryos, so he was losing faith in the effort. “You see many failed attempts and get disappointed,” he says.
Gambini and his team, a group of Argentinian and Spanish researchers, were working to create these embryos because many donkey populations around the world are declining. It may sound counterintuitive that domesticated animals may need preservation, but out of 28 European donkey breeds, 20 are endangered and seven are in critical status. It is partly because of the inbreeding that happened over the course of many years and partly because in today’s Western world donkeys aren’t really used anymore.
“That's the reason why some breeds begin to disappear because humans were not really interested in having that specific breed anymore,” Gambini says. Nonetheless, in Africa, India and Latin America millions of rural families still rely on these hardy creatures for agriculture and transportation. And the only two wild donkey species—Equus africanus in Africa and Equus hemionus in Asia—are also dwindling, due to losing their habitats to human activities, diseases and slow reproduction rates. Gambini’s team wanted to create a way to preserve the animals for the future. “Donkeys are more endangered than people realize,” he says.
There’s much more to donkeys' trouble though. For the past 20 or so years, they have been facing a huge existential threat due to their hide gelatin, a compound derived from their skins by soaking and stewing. In Chinese traditional medicine, the compound, called ejiao, is believed to have a medicinal value, so it’s used in skin creams, added to food and taken in capsules. Centuries ago, ejiao was a very expensive luxury product available only for the emperor and his household. That changed in the 1990s when the Chinese economy boomed, and many people were suddenly able to afford it. “It went from a very elite product to a very popular product,” says Janneke Merkx, a campaign manager at The Donkey Sanctuary, a United Kingdom-based nonprofit organization that keeps tabs on the animals’ welfare worldwide. “It is a status symbol for gift giving.”
Having evolved in the harsh and arid mountainous terrains where food and water were scarce, donkeys are extremely adaptable and hardy. But the Donkey Sanctuary documented cases in which an entire village had their animals disappear overnight, finding them killed and skinned outside their settlement.
The Chinese donkey population was quickly decimated. Unlike many other farm animals, donkeys are finicky breeders. When stressed and unhappy, they don’t procreate, so growing them in large industrial settings isn’t possible. “Donkeys are notoriously slow breeders and really very difficult to farm,” says Merkx. “They are not the same as other livestock like sheep and pigs and cattle.” Within years the, the donkey numbers in China dropped precipitously. “China used to have the largest donkey population in the world in the 1990s. They had 11 million donkeys, and it's now down to less than 3 million, and they just can't keep up with the demand.”
To keep the ejiao conveyor going, some producers turned to the illegal wildlife trade. Poachers began to steal and slaughter donkeys from rural villages in Africa. The Donkey Sanctuary documented cases in which an entire village had their animals disappear overnight, finding them killed and skinned outside their settlement. Exactly how many creatures were lost to the skin trade to-date isn’t possible to calculate, says Faith Burden, the Donkey Sanctuary’s director of equine operations. Traditionally a poor people’s beast of burden, donkey counts are hard to keep track of. “When an animal doesn't produce meat, milk or eggs or whatever edible product, they're often less likely to be acknowledged in a government population census,” Burden says. “So reliable statistics are hard to come by.” The nonprofit estimates that about 4.8 million are slaughtered annually.
During their six to seven thousand years of domestication, donkeys rarely got the full appreciation for their services. They are often compared to horses, which doesn’t do them justice. They’re entirely different animals, Burden says. Built for speed, horses respond to predators and other dangers by running as fast as they can. Donkeys, which originate from the rocky, mountainous regions of Africa where running is dangerous, react to threats by freezing and assessing the situation for the best response. “Those so-called stubborn donkeys that won’t move as you want, they are actually thinking ‘what’s the best approach,’” Burden says. They may even choose to fight the predators rather than flee, she adds. “In some parts of the world, people use them as guard animals against things like coyotes and wolves.”
Scientists believe that domestic donkeys take their origin from Equus africanus or African wild ass, originally roaming where Kenya, Ethiopia and Eritrea are today. Having evolved in the harsh and arid mountainous terrains where food and water were scarce, they are extremely adaptable and hardy. Research finds that they can go without water for 72 hours and then drink their fill without any negative consequences. Their big jaws let them chew tough desert shrubs, which horses can’t exist on. Their large ears help dissipate heat. Their little upright hooves are a perfect fit for the uneven rocky or other dangerous grounds. Accustomed to the mountain desert climate with hot days and cold nights, they don’t mind temperature flux.
“The donkey is the most supremely adapted animal to deal with hostile conditions,” Burden says. “They can survive on much lower nutritional quality food than a cow, sheep or horse. That’s why communities living in some of the most inhospitable places will often have donkeys with them.” And that’s why losing a donkey to an illegal skin trade can devastate a family in places like Eritrea. Suddenly everything from water to firewood to produce must be carried by family members—and often women.

Workers unloading donkeys at the Shinyanga slaughterhouse in Tanzania. Fearing a future in which donkeys go extinct, scientists have found ways to cryopreserve a donkey embryo in liquid nitrogen.
TAHUCHA
One can imagine a time when worldwide donkey populations may dwindle to the point that they would need to be restored. That includes their genetic variability too. That’s where the frozen embryos may come in handy. We may be able to use them to increase the genetic variability of donkeys, which will be especially important if they get closer to extinction, Gambini says. His team had already created frozen embryos for horses and zebras, an idea similar to a seed bank. “We call this concept the Frozen Zoo.”
Creating donkey embryos proved much harder than those of zebras and horses. To improve chances of fertilization, Gambini used the intracytoplasmic sperm injection or ICSI, in which he employed a tiny needle called a micropipette to inject a donkey sperm into an egg. That was a step above the traditional IVF method, in which the egg and a sperm are left floating in a test tube together. The injection took, but during the incubating week, one after the other, the embryos stopped dividing. Finally, on day seven, Gambini finally spotted the exact sight he was hoping to see. One of the embryos developed into a burgeoning ball of cells.
“That stage is called a blastocyst,” Gambini says. The clump of cells had a lot of fluids mixed within them, which indicated that they were finally developing into a viable embryo. “When we see a blastocyst, we know we can transfer that into a female.” He was so excited he immediately called all his collaborators to tell them the good news, which they later published in the journal of Theriogenology.
The one and only embryo to reach that stage, the blastocyst was cryopreserved in liquid nitrogen. The team is waiting for the next breeding season to see if a female donkey may carry it to term and give birth to a healthy foal. Gambini’s team is hoping to polish the process and create more embryos. “It’s our weapon in the conservation ass-enal,” he says.
Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.
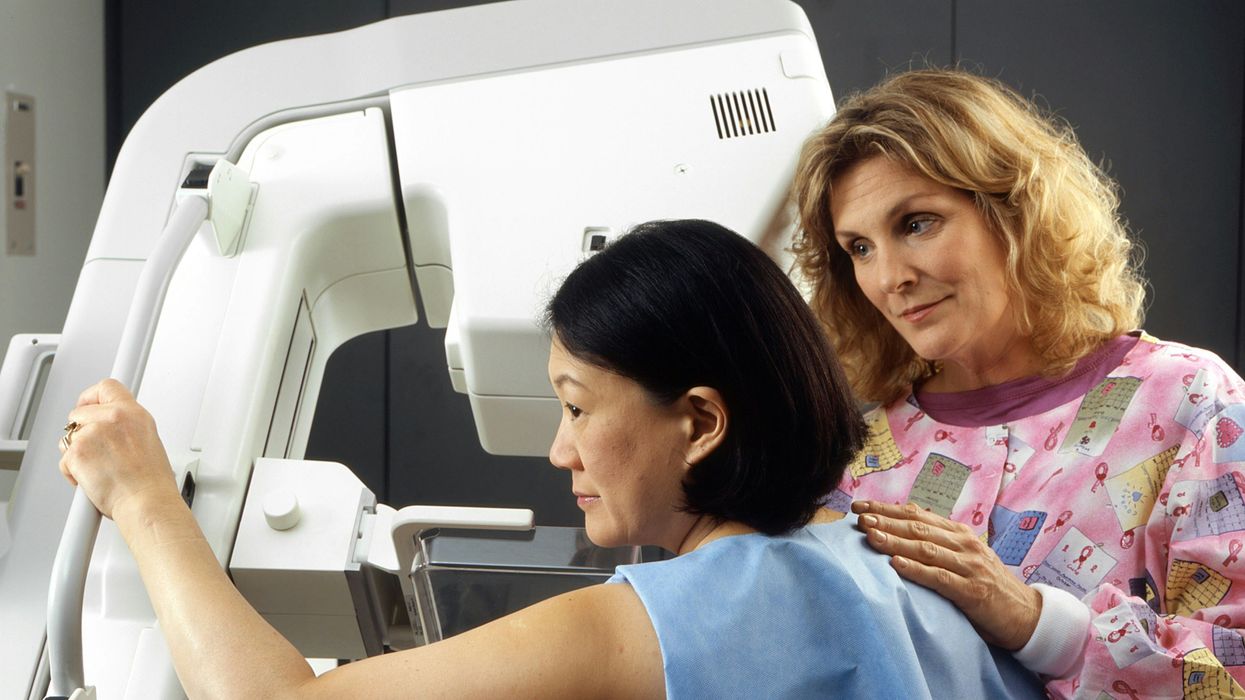
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.
Urinary tract infections account for more than 8 million trips to the doctor each year.
Few things are more painful than a urinary tract infection (UTI). Common in men and women, these infections account for more than 8 million trips to the doctor each year and can cause an array of uncomfortable symptoms, from a burning feeling during urination to fever, vomiting, and chills. For an unlucky few, UTIs can be chronic—meaning that, despite treatment, they just keep coming back.
But new research, presented at the European Association of Urology (EAU) Congress in Paris this week, brings some hope to people who suffer from UTIs.
Clinicians from the Royal Berkshire Hospital presented the results of a long-term, nine-year clinical trial where 89 men and women who suffered from recurrent UTIs were given an oral vaccine called MV140, designed to prevent the infections. Every day for three months, the participants were given two sprays of the vaccine (flavored to taste like pineapple) and then followed over the course of nine years. Clinicians analyzed medical records and asked the study participants about symptoms to check whether any experienced UTIs or had any adverse reactions from taking the vaccine.
The results showed that across nine years, 48 of the participants (about 54%) remained completely infection-free. On average, the study participants remained infection free for 54.7 months—four and a half years.
“While we need to be pragmatic, this vaccine is a potential breakthrough in preventing UTIs and could offer a safe and effective alternative to conventional treatments,” said Gernot Bonita, Professor of Urology at the Alta Bro Medical Centre for Urology in Switzerland, who is also the EAU Chairman of Guidelines on Urological Infections.
The news comes as a relief not only for people who suffer chronic UTIs, but also to doctors who have seen an uptick in antibiotic-resistant UTIs in the past several years. Because UTIs usually require antibiotics, patients run the risk of developing a resistance to the antibiotics, making infections more difficult to treat. A preventative vaccine could mean less infections, less antibiotics, and less drug resistance overall.
“Many of our participants told us that having the vaccine restored their quality of life,” said Dr. Bob Yang, Consultant Urologist at the Royal Berkshire NHS Foundation Trust, who helped lead the research. “While we’re yet to look at the effect of this vaccine in different patient groups, this follow-up data suggests it could be a game-changer for UTI prevention if it’s offered widely, reducing the need for antibiotic treatments.”