Do-It-Yourself Diabetes Systems Bring Convenience—And Risk

Dana Lewis, pictured in Mount Vernon in 2017, worked with her engineer husband to design an artificial pancreas system to manage her type 1 diabetes.
For years, a continuous glucose monitor would beep at night if Dana Lewis' blood sugar measured too high or too low. At age 14, she was diagnosed with type 1 diabetes, an autoimmune disease that destroys insulin-producing cells in the pancreas.
The FDA just issued its first warning to the DIY diabetic community, after one patient suffered an accidental insulin overdose.
But being a sound sleeper, the Seattle-based independent researcher, now 30, feared not waking up. That concerned her most when she would run, after which her glucose dropped overnight. Now, she rarely needs a rousing reminder to alert her to out-of-range blood glucose levels.
That's because Lewis and her husband, Scott Leibrand, a network engineer, developed an artificial pancreas system—an algorithm that calculates adjustments to insulin delivery based on data from the continuous glucose monitor and her insulin pump. When the monitor gives a reading, she no longer needs to press a button. The algorithm tells the pump how much insulin to release while she's sleeping.
"Most of the time, it's preventing the frequent occurrences of high or low blood sugars automatically," Lewis explains.
Like other do-it-yourself device innovations, home-designed artificial pancreas systems are not approved by the Food and Drug Administration, so individual users assume any associated risks. Experts recommend that patients consult their doctor before adopting a new self-monitoring approach and to keep the clinician apprised of their progress.
DIY closed-loop systems can be uniquely challenging, according to the FDA. Patients may not fully comprehend how the devices are intended to work or they may fail to recognize the limitations. The systems have not been evaluated under quality control measures and pose risks of inappropriate dosing from the automated algorithm or potential incompatibility with a patient's other medications, says Stephanie Caccomo, an FDA spokeswoman.
Earlier this month, in fact, the FDA issued its first warning to the DIY diabetic community, which includes thousands of users, after one patient suffered an accidental insulin overdose.
Patients who built their own systems from scratch may be more well-versed in the operations, while those who are implementing unapproved designs created by others are less likely to be familiar with their intricacies, she says.
"Malfunctions or misuse of automated-insulin delivery systems can lead to acute complications of hypo- and hyperglycemia that may result in serious injury or death," Caccomo cautions. "FDA provides independent review of complex systems to assess the safety of these nontransparent devices, so that users do not have to be software/hardware designers to get the medical devices they need."
Only one hybrid closed-loop technology—the MiniMed 670G System from Minneapolis-based Medtronic—has been FDA-approved for type 1 use since September 2016. The term "hybrid" indicates that the system is not a fully automatic closed loop; it still requires minimal input from patients, including the need to enter mealtime carbohydrates, manage insulin dosage recommendations, and periodically calibrate the sensor.
Meanwhile, some tech-savvy people with type 1 diabetes have opted to design their own systems. About one-third of the DIY diabetes loopers are children whose parents have built them a closed system, according to Lewis' website.
Lewis began developing her system in 2014, well before Medtronic's device hit the market. "The choice to wait is not a luxury," she says, noting that "diabetes is inherently dangerous," whether an individual relies on a device to inject insulin or administers it with a syringe.
Hybrid closed-loop insulin delivery improves glucose control while decreasing the risk of low blood sugar in patients of various ages with less than optimally controlled type 1 diabetes, according to a study published in The Lancet last October. The multi-center randomized trial, conducted in the United Kingdom and the United States, spanned 12 weeks and included adults, adolescents, and children aged 6 years and older.
"We have compelling data attesting to the benefits of closed-loop systems," says Daniel Finan, research director at JDRF (formerly the Juvenile Diabetes Research Foundation) in New York, a global organization funding the study.
Medtronic's system costs between $6,000 and $9,000. However, end-user pricing varies based on an individual's health plan. It is covered by most insurers, according to the device manufacturer.
To give users more choice, in 2017 JDRF launched the Open Protocol Automated Insulin Delivery Systems initiative to collaborate with the FDA and experts in the do-it-yourself arena. The organization hopes to "forge a new regulatory paradigm," Finan says.
As diabetes management becomes more user-controlled, there is a need for better coordination. "We've had insulin pumps for a very long time, but having sensors that can detect blood sugars in real time is still a very new phenomenon," says Leslie Lam, interim chief in the division of pediatric endocrinology and diabetes at The Children's Hospital at Montefiore in the Bronx, N.Y.
"There's a lag in the integration of this technology," he adds. Innovators are indeed working to bring new products to market, "but on the consumer side, people want that to be here now instead of a year or two later."
The devices aren't foolproof, and mishaps can occur even with very accurate systems. For this reason, there is some reluctance to advocate for universal use in children with type 1 diabetes. Supervision by a parent, school nurse, and sometimes a coach would be a prudent precaution, Lam says.
People engage in "this work because they are either curious about it themselves or not getting the care they need from the health care system, or both."
Remaining aware of blood sugar levels and having a backup plan are essential. "People still need to know how to give injections the old-school way," he says.
To ensure readings are correct on Medtronic's device, users should check their blood sugar with traditional finger pricking at least five or six times per day—before every meal and whenever directed by the system, notes Elena Toschi, an endocrinologist and director of the Young Adult Clinic at Joslin Diabetes Center, an affiliate of Harvard Medical School.
"There can be pump failure and cross-talking failure," she cautions, urging patients not to stop being vigilant because they are using an automated device. "This is still something that can happen; it doesn't eliminate that."
While do-it-yourself devices help promote autonomy and offer convenience, the lack of clinical trial data makes it difficult for clinicians and patients to assess risks versus benefits, says Lisa Eckenwiler, an associate professor in the departments of philosophy and health administration and policy at George Mason University in Fairfax, Va.
"What are the responsibilities of physicians in that context to advise patients?" she questions. Some clinicians foresee the possibility that "down the road, if things go awry" with disease management, that could place them "in a moral quandary."
Whether it's controlling diabetes, obesity, heart disease or asthma, emerging technologies are having a major influence on individuals' abilities to stay on top of their health, says Camille Nebeker, an assistant professor in the School of Medicine at the University of California, San Diego, and founder and director of its Research Center for Optimal Data Ethics.
People engage in "this work because they are either curious about it themselves or not getting the care they need from the health care system, or both," she says. In "citizen science communities," they may partner in participant-led research while gaining access to scientific and technical expertise. Others "may go it alone in solo self-tracking studies or developing do-it-yourself technologies," which raises concerns about whether they are carefully considering potential risks and weighing them against possible benefits.
Dana Lewis admits that "using do-it-yourself systems might not be for everyone. But the advances made in the do-it-yourself community show what's possible for future commercial developments, and give a lot of hope for improved quality of life for those of us living with type 1 diabetes."
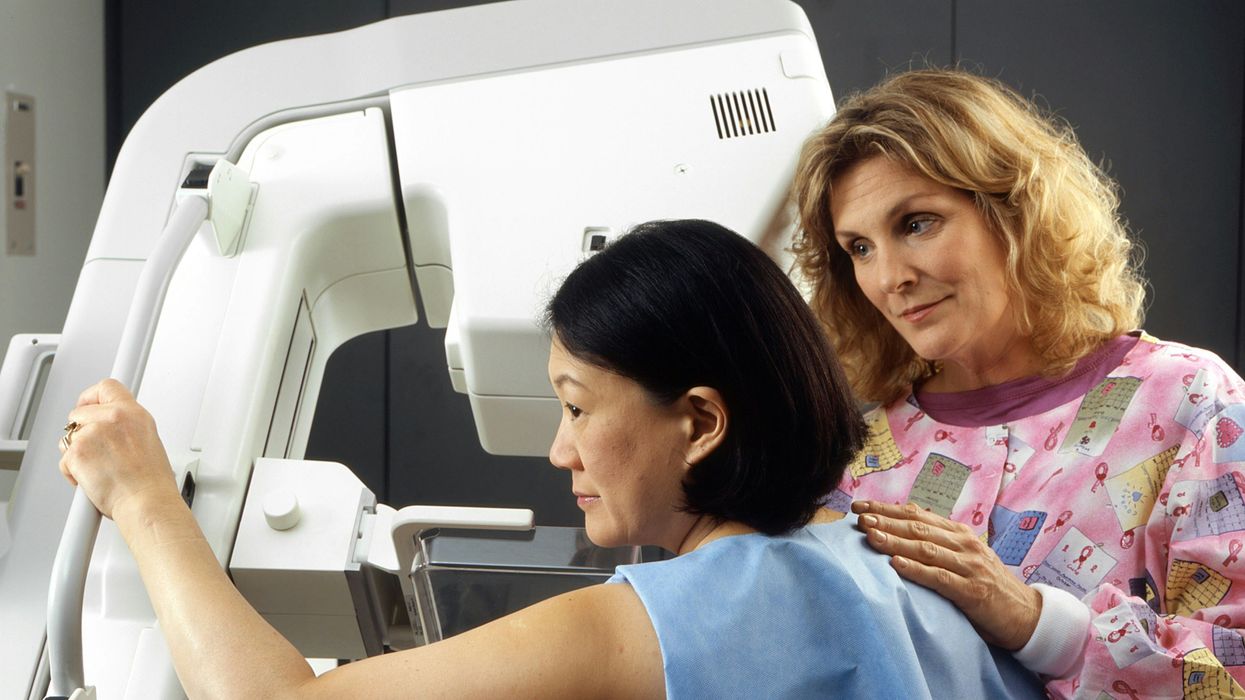
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.
Urinary tract infections account for more than 8 million trips to the doctor each year.
Few things are more painful than a urinary tract infection (UTI). Common in men and women, these infections account for more than 8 million trips to the doctor each year and can cause an array of uncomfortable symptoms, from a burning feeling during urination to fever, vomiting, and chills. For an unlucky few, UTIs can be chronic—meaning that, despite treatment, they just keep coming back.
But new research, presented at the European Association of Urology (EAU) Congress in Paris this week, brings some hope to people who suffer from UTIs.
Clinicians from the Royal Berkshire Hospital presented the results of a long-term, nine-year clinical trial where 89 men and women who suffered from recurrent UTIs were given an oral vaccine called MV140, designed to prevent the infections. Every day for three months, the participants were given two sprays of the vaccine (flavored to taste like pineapple) and then followed over the course of nine years. Clinicians analyzed medical records and asked the study participants about symptoms to check whether any experienced UTIs or had any adverse reactions from taking the vaccine.
The results showed that across nine years, 48 of the participants (about 54%) remained completely infection-free. On average, the study participants remained infection free for 54.7 months—four and a half years.
“While we need to be pragmatic, this vaccine is a potential breakthrough in preventing UTIs and could offer a safe and effective alternative to conventional treatments,” said Gernot Bonita, Professor of Urology at the Alta Bro Medical Centre for Urology in Switzerland, who is also the EAU Chairman of Guidelines on Urological Infections.
The news comes as a relief not only for people who suffer chronic UTIs, but also to doctors who have seen an uptick in antibiotic-resistant UTIs in the past several years. Because UTIs usually require antibiotics, patients run the risk of developing a resistance to the antibiotics, making infections more difficult to treat. A preventative vaccine could mean less infections, less antibiotics, and less drug resistance overall.
“Many of our participants told us that having the vaccine restored their quality of life,” said Dr. Bob Yang, Consultant Urologist at the Royal Berkshire NHS Foundation Trust, who helped lead the research. “While we’re yet to look at the effect of this vaccine in different patient groups, this follow-up data suggests it could be a game-changer for UTI prevention if it’s offered widely, reducing the need for antibiotic treatments.”