We have a small favor to ask of you
Facebook is critical to our success and we could use your help. It will only take a few clicks on your device. But it would mean the world to us.
Here’s the link . Once there, hit the Follow button. Hit the Follow button again and choose Favorites. That’s it!
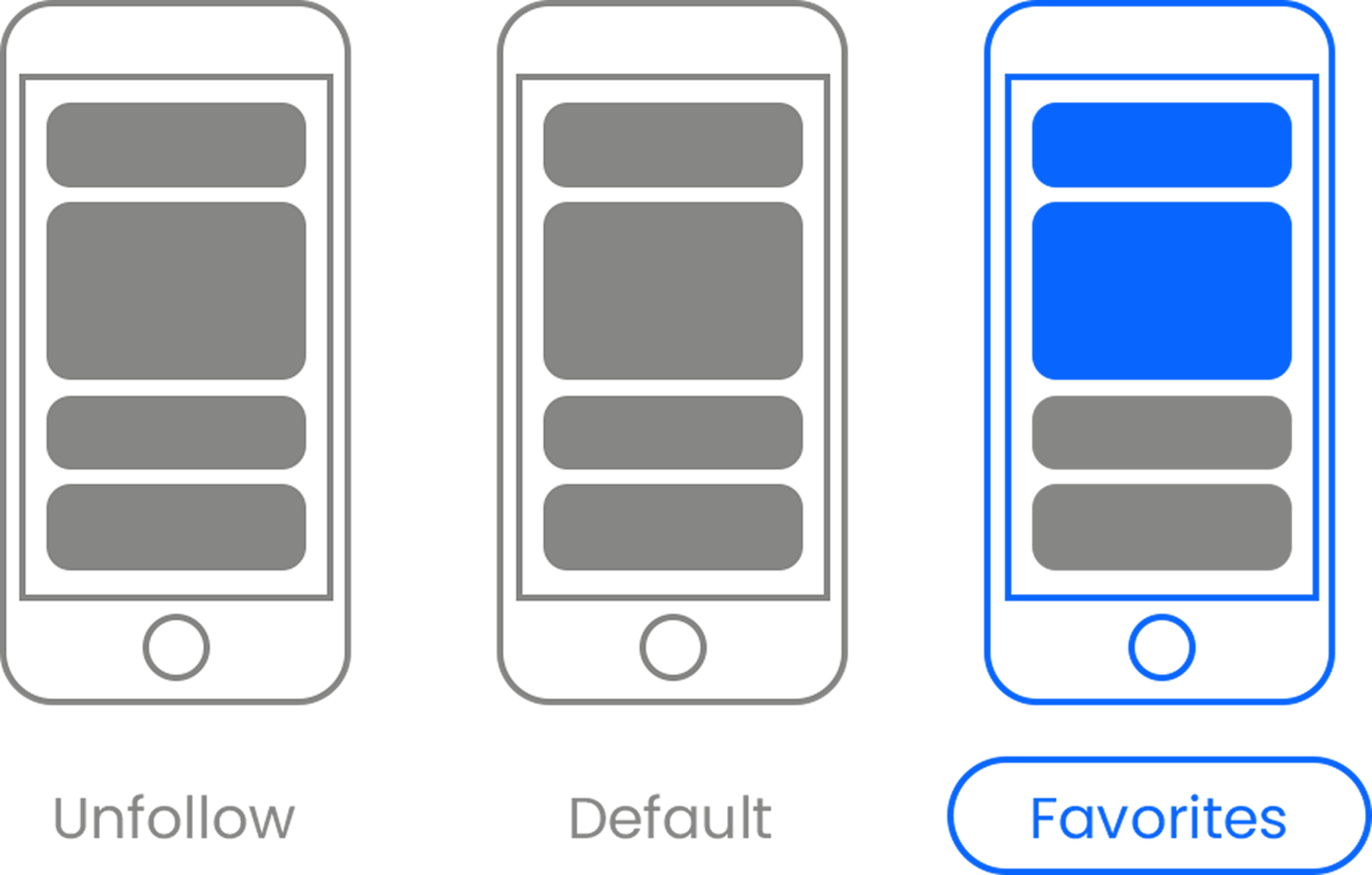
If you’d like to know why this is so important for us, you can read more about it here .



